The time-dependent Schrödinger equation (TDSE) describes the quantum dynamical nature of molecular processes. Simulations are, however, computationally very demanding due to the curse of dimensionality. With our MPI and OpenMP parallelized code, HAParaNDA (cf.[1]), we are able to accurately solve the full Schrödinger equation, currently in up to five dimensions using a medium-size cluster.
The ![]() -dimensional TDSE reads
-dimensional TDSE reads
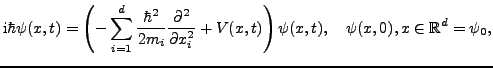 |
In order to demonstrate the performance of a massively parallel
simulation of the TDSE based on a FD-Lanczos discretization, we have
conducted several simulations on a medium-size cluster, consisting of 316
nodes. Each node is equipped with dual quad-core Intel Nehalem CPU and 24
GB of DRAM. The nodes are interconnected by an InfiniBand fabric. For the
experiments, we have considered the harmonic oscillator and the
Henon-Heiles potential. The analytical solution is known for the
harmonic oscillator, so we are able to verify the correctness and the
accuracy of our numerical results. Table ![]() shows the
scalability of the results for the 4D harmonic oscillator. Comparing the
variants of the Lanczos algorithm, we see that the performance can indeed
be improved by reducing the communication. We are currently working on an
implementation of the
shows the
scalability of the results for the 4D harmonic oscillator. Comparing the
variants of the Lanczos algorithm, we see that the performance can indeed
be improved by reducing the communication. We are currently working on an
implementation of the ![]() -step method and hope to further improve the
scalability in that way. The Henon-Heiles potential is a common test
case for high-dimensional simulations. We have simulated this problem in
five dimensions on a grid with
-step method and hope to further improve the
scalability in that way. The Henon-Heiles potential is a common test
case for high-dimensional simulations. We have simulated this problem in
five dimensions on a grid with ![]() points over
points over ![]() time steps. We
used the standard Lanczos algorithm and have chosen the size of the
Krylov space adaptively. The simulation was performed on 1024 cores in 19
hours.
time steps. We
used the standard Lanczos algorithm and have chosen the size of the
Krylov space adaptively. The simulation was performed on 1024 cores in 19
hours.
| # Cores | 8 | 16 | 32 | 64 | 128 | 256 | 512 |
| L1 | 3.92 | 4.31 | 4.37 | 4.55 | 4.72 | 4.87 | 5.16 |
| L2 | 3.79 | 4.06 | 4.26 | 4.33 | 4.43 | 4.72 | 4.83 |
| [1] | M. Gustafsson and S. Holmgren. An implementation framework for solving high-dimensional PDEs on massively parallel computers, to appear in: Proceedings of ENUMATH 2009, Uppsala, Sweden |
| [2] | S.K. Kim and A.T. Chronopoulos. A class of Lanczos-like algorithms implemented on parallel computers, Parallel Comput. 17 (1991) |
| [3] | K. Kormann, S. Holmgren, and H.O. Karlsson. Accurate time propagation for the Schrödinger equation with an explicitly time-dependent Hamiltonian, J. Chem. Phys. 128 (2008) |
| [4] | K. Kormann and A. Nissen. Error Control for Simulations of a Dissociative Quantum System, to appear in: Proceedings of ENUMATH 2009, Uppsala, Sweden |